Stanford researchers conduct census of cell surface proteins
A new technique for systematically surveying proteins on the outer surface of cells, which act like molecular social cues to guide cell-cell interactions and assembly into tissues and organs.
Stanford scientists have completed the first global census of diverse proteins sprouting tree-like from the outer membrane of a cell. These cell surface proteins govern how cells interact with one another and how they assemble into tissues and organs such as hearts and brains.
“Cell architecture is determined by cell-cell interactions, and these interactions are mediated by molecules on the cell surface. They’re the business end of things,” said Jiefu Li, a graduate student in the biology department at Stanford University.
Knowing what proteins are embedded on cell surfaces and what they do is a first step toward understanding how individual cells fashion themselves into intricate structures like organs—and how to prevent or treat diseases that occur when these molecular social cues are absent or misread.
“Our brains are made up of 100 billion neurons that make 100 trillion synaptic connections,” said Liqun Luo, the Ann and Bill Swindells Professor in the School of Humanities and Sciences at Stanford. “How do brain cells know who to make connections with during development? That’s a fundamental question in neurobiology.”
Luo, Li, and their colleagues took an important new step toward answering this question in a study published online on January 16 in the journal Cell. In it the scientists report surveying cell surface proteins on a specific type of brain cell in fruit flies, whose neurons closely mimic our own.
By comparing the brains of young and adult flies, the scientists showed how the population of proteins changes over time as the insects mature and the needs of the cells change.
An agnostic approach
A coauthor on the study, Alice Ting, a professor of biology and genetics, said the big difference between this approach and previous efforts to understand cell surface proteins lies in the fact that the group didn’t know what they were looking for.
In the past, the search for new cell surface proteins was heavily influenced by proteins that had been discovered before. If a cell surface protein was known to help similar neurons connect with one another, for example, scientists would explore other proteins in the same molecular family to see whether they had similar properties. This approach worked, but it limited the likelihood of encountering completely new cell surface proteins.
“What's cool here is that we're not looking only for proteins that resemble known cell surface proteins with the functions that we're interested in,” said Ting, who is also a Chan Zuckerberg Biohub investigator. “We can go in without any prior assumptions and just see what turns up.”
This powerful agnostic approach was made possible through “proximity labeling,” a technique pioneered by Ting’s group that involves genetically engineering proteins to mark their closest neighbors with a unique molecular tag.
“It’s like spray painting,” Ting said. “When spray painting, the paint distribution is going to be densest in the regions closest to your paint bottle.”
Biotin
For the new study, Luo’s lab employed proximity labeling to mark all of the cell surface proteins on a particular group of neurons in the brains of developing and adult fruit flies.
The scientists used a special strain of flies bred to produce a catalytic protein, or enzyme, borrowed from horseradish plants. This enzyme, called peroxidase, functions like an unopened can of spray paint and only resides on the surface of a particular kind of neuron in the fly brains that the scientists wanted to study.
At different points in the flies’ development, the scientists removed the brains and did the molecular equivalent of triggering the spray cans to paint all the nearby proteins. This involved immersing the cells in chemical baths that caused the horseradish proteins to mark their cell surface neighbors with a molecule called biotin.
Next, the researchers broke apart the fly brains and their constituent neurons, and pulled out all the proteins that got sprayed – or, rather, that contained the biotin tag. “The beauty here is that you’re only enriching the cell surface proteins and ignoring all of the proteins inside the cell,” said Luo, who is also an investigator at the Howard Hughes Medical Institute.
This study was the first time the researchers used the technique in a living organism rather than in a petri dish. “To get this to actually work in vivo”—in intact brains of live organisms—“was quite a technical leap,” Ting said.
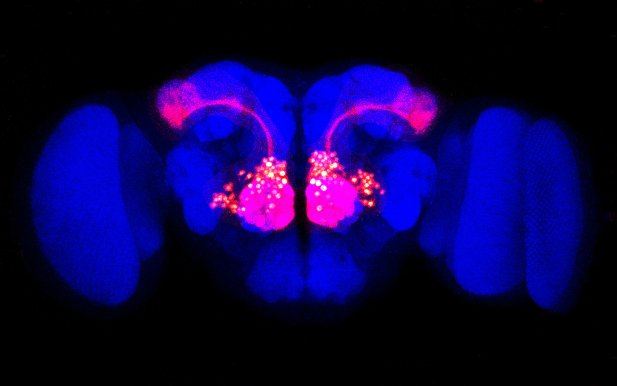
Olfactory projection neurons (red) are visible against the outline of a Drosophila fly brain (blue).
Beyond the lamppost
In relatively few steps, the scientists were able to conduct a complete census of the more than 700 different kinds of protein that dot the surface of olfactory projection neurons in fruit flies. The team’s haul included many cell surface proteins that had been painstakingly identified previously using other techniques.
Even more intriguing, their survey also turned up 20 new cell surface proteins in the brains of the developing flies, which subsequent experiments by the team revealed to be important for brain wiring.
“This shows that this approach is really unbiased,” Ting said. “It allows you to look away from under the lamppost to find proteins that you may miss otherwise.”
To read all stories about Stanford science, subscribe to the biweekly Stanford Science Digest.
Other Stanford coauthors include graduate students Shuo Han (a co-first author on the study and a Stanford Bio-X Bowes Fellow), Pornchai Kaewsapsak, Chuanyun Xu, Qijing Xie, and Bing Wu; postdoctoral scholars Hongjie Li, Tongchao Li, and Colleen McLaughlin; undergraduates Ricardo Guajardo (a Bio-X undergraduate fellow) and Anthony Xie; research associate David Luginbuhl; and Stephen Quake, the Lee Otterson Professor in the School of Engineering, Bioengineering, and Applied Physics. Researchers at the Broad Institute of MIT and Harvard also contributed to this study.
Luo, Ting, and Quake are members of Stanford Bio-X, the Wu Tsai Neurosciences Institute, and the Stanford Cancer Institute. Luo and Ting are also faculty fellows in Stanford ChEM-H.
The research was supported by the National Institutes of Health and is part of a “Neuro-omics” proposal funded by the Wu Tsai Neurosciences Institute at Stanford University that aims to advance the understanding of the neural basis of behavior by developing and disseminating methods for studying the genome, proteins and neural networks at work in the brain.