Study uncovers unique patterns of native vascular plant diversity
A new study by Stanford biologist Barnabas Daru uses his previous research that identified coverage gaps and biases in biodiversity data to create more accurate estimates of plant distribution for over 200,000 native plant species worldwide.
It is hard to protect something if you don’t know where it is. Yet many people who study and want to safeguard native plants are faced with this exact problem.
There are roughly 340,000 species of plants with water transporting tissues, called vascular plants. People are most familiar with a tiny subset of vascular plants, such as trees, agricultural crops, and flowering plants for the products, food, and beauty they provide. Yet all vascular plants play important roles in maintaining ecosystem processes and supporting and feeding life on Earth.
Now, a new Stanford study that used machine learning techniques to overcome biases in biodiversity data fills in the once patchy global map of vascular plant distributions. The study revealed previously unknown patterns of native vascular plant diversity and found that approximately 60% of plant diversity hotspots are located outside of protected areas.
“The entire biosphere depends on plants,” said Barnabas Daru, assistant professor of biology in the Stanford School of Humanities and Sciences. “But if we don't know their distributions it is challenging to know how they are doing or if they are being threatened by climate change or human activities.”
The findings of Daru’s research were published August 12 in the Proceedings of the National Academies of Sciences.
Making biased data better
Most records of plant diversity are in the form of physical specimens in herbaria (museums for plants) or digital field observations. These data are useful, but as Daru found in his 2023 study published in Nature Ecology & Evolution, they contain widespread biases and coverage gaps.
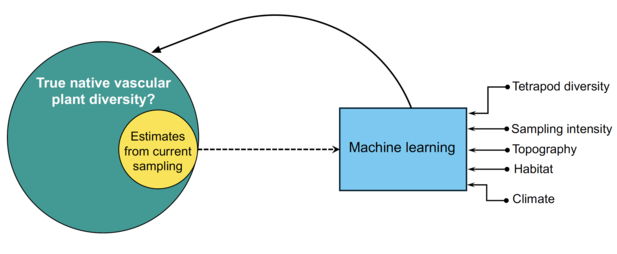
Researchers often use species distribution models to predict where species ought to be. These models use data on the occurrence and abundance of each species, combined with environmental variables that affect their survival, such as temperature and rainfall.
“The problem with this approach is that the input biodiversity data are already biased,” Daru said. “So we are likely to get biased predictions for species distributions.”
Daru’s method uses existing biodiversity data and environmental variables—as traditional approaches have done before to obtain modeled estimates—but with additional data inputs and modifications to make the model’s predictions more accurate.
“My approach incorporates maps of sampling biases” Daru explained. “This trains the modeled estimates using a machine learning model as a function of the biased nature of the data and other factors that determine plant distributions. If certain locations are oversampled and other regions are undersampled, the model can account for the uneven sampling biases. Then I added another layer and incorporated the dispersal rates of the different plant species.”
Traditional species distribution models predict where species are likely to be found based on the suitability of different climates for each species, Daru explained. But just because a certain climate is suitable for a particular species, doesn’t mean the species will be found there.
“The South African native ice plant (Carpobrotus edulis) and California poppy (Eschscholzia californica) are good examples,” Daru said. “If we use a species distribution model to predict the niches of the ice plant it will show that ice plants can find suitable habitat in the South African Cape, California and other Mediterranean-type regions with similar climates.”
Similarly, species distribution models predict that the California poppy should be found in the South African Cape and other biomes with similar climates, but the poppy is native to only mediterranean California.
“If your model doesn’t account for the dispersal rates of each species—that ice plants and California poppies cannot cross the oceans to populate the other hemisphere on their own without human help in the form of invasive species introductions—you will have inaccurate species distribution model predictions,” Daru said.
Daru obtained the dispersal rates for more than 200,000 different species of vascular plants using spherical Brownian motion models to determine the rate each species can disperse based on information known about its evolutionary history and the conditions it needs to survive.
As a final input into his modified model, Daru included data on the distribution of well-studied birds, mammals, amphibians, and reptiles because their geographic sampling is more accurate and these organisms often live near vascular plants.
One reason previous studies have not attempted to include dispersal rates—or other factors that could improve accuracy—in their plant distribution calculations is that most tools cannot handle massive datasets at a global scale, Daru explained.
“The framework for calculating dispersal rates was developed for microorganisms that disperse much shorter distances than plants,” Daru said. “That was one challenge. Another challenge was the computational part—there are multiple steps involved in building the distribution maps that generate a lot of data.”
Daru ran the model about five times for each species. Then he divided a map of the globe into pixels representing 20 by 20 kilometer plots and computed the number of species within each pixel.
“The final matrix was massive, but my lab develops the bioinformatics tools that enable us to handle massive datasets,” Daru said.
Protecting plant diversity hotspots
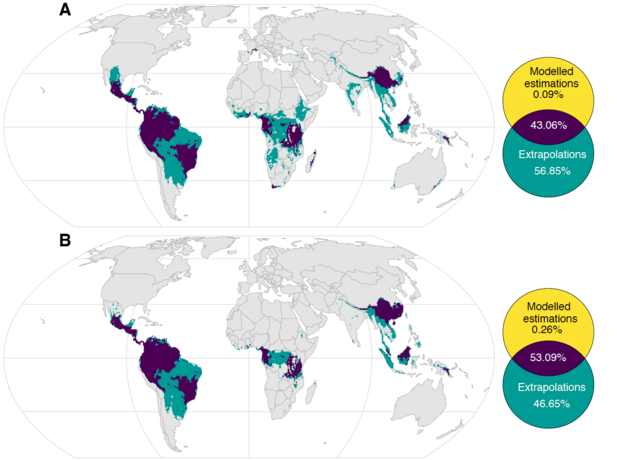
The resulting maps revealed clusters of vascular plant species richness in known biodiversity hotspots, like the Amazon and Madagascar, but also in unexpected places like Chaco, Argentina; the Cerrado savannas, South America; the Democratic Republic of the Congo; and Yunnan, China. He also found that places with high native species richness also had high phylogenetic (evolutionary) diversity, and both species richness and phylogenetic (evolutionary) diversity were greatest near the equator and lower at higher latitudes.
Daru tested if these plant diversity hotspots are captured within the borders of game reserves, national parks, and other protected areas. He found that most facets of plant diversity are unprotected and approximately 60% of vascular plant diversity lies outside of protected areas.
“If these plants are not protected, then all the organisms that depend on them are equally not protected,” Daru said.
Daru also found that trees and other large plant species are often sheltered within protected areas, but evolutionary distinctive plants with few or no close living relatives are not.
“If we lose these unique plant species that would be a huge loss to the evolutionary history of plants,” Daru said. “Suggesting that, yes, it is indeed worth expanding protected areas to include evolutionary distinctive plants and other attributes of plant diversity.”
In the future, Daru would like to develop a mobile app that tells users the number and species of plants within a given radius. As users validate whether the predicted plants are present, the app could help researchers improve the quality of biodiversity data collected in the future.
“We know a lot about birds, mammals, and other charismatic animals, but we don't know as much about plants or their global distributions,” Daru said. “This study’s findings can advance our knowledge of plant ecology and biodiversity in ways that were not possible before and—for the first time—can help us prioritize areas for plant conservation.”
Acknowledgements
This research was supported by the U.S. National Science Foundation.
To read all stories about Stanford science, subscribe to the biweekly Stanford Science Digest.
Media contact
Joy Leighton, Stanford School of Humanities and Sciences: joy [dot] leighton [at] stanford [dot] edu (joy[dot]leighton[at]stanford[dot]edu)